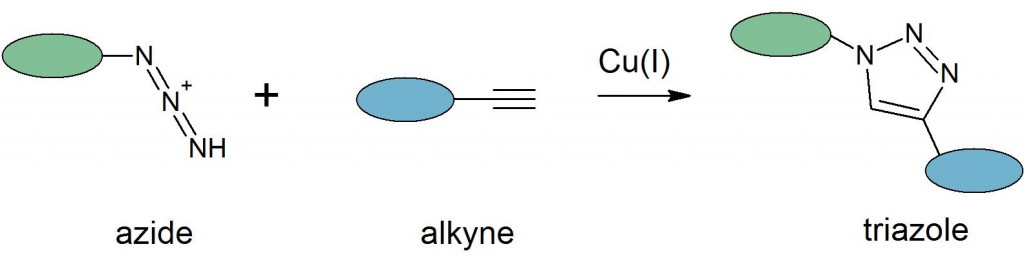
Click chemistry provides straightforward approaches for selectively labeling or linking biomolecules. Traditional bioconjugations often can’t be carried out within complex samples or under physiological conditions and lack specificity for site-specific labeling. The development of click chemistry approaches has led to the use of pairs of ‘bioorthogonal’ reactive groups that are inert to endogenous biomolecules and that selectively react under mild, aqueous conditions with high efficiency. The prototypical click reaction for ligating or labeling biomolecules is the azide-alkyne cycloaddition:
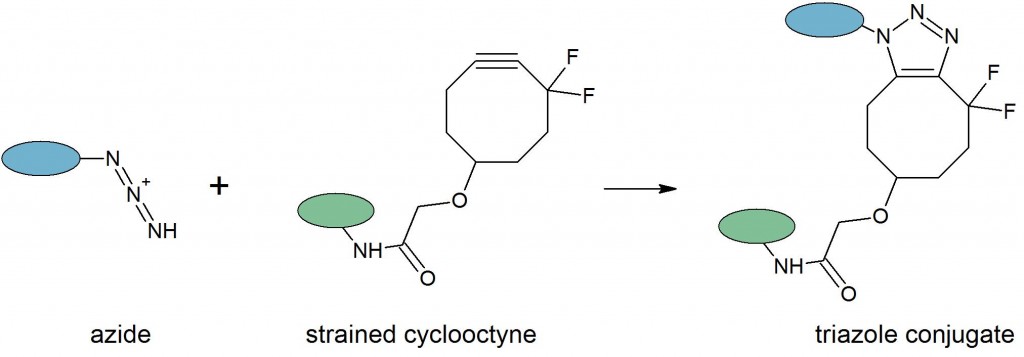
This reaction offers the advantages of being rapid, highly selective, and compatible with aqueous buffers. Yields are typically high, eliminating the need for purification of the product. In addition, the small size of the reactants and the triazole product create only minor structural changes in the labeled molecule that are accepted by many enzyme or protein binding partners. The azide-alkyne cycloaddition is typically catalyzed through proximity, a copper(I) catalyst or in copper-free reactions, driven by relieving the ring strain of a cyclooctyne:
The advantages provided by click chemistry facilitate a wide range of applications such as specific affinity or fluorescent labeling, linking molecules to solid matrices as bait for affinity purifications or ligand fishing, and opens the door to cell-based or in vivo labeling of targets. Below are just a few examples of how click chemistry can be applied to labeling biomolecules, cell-based experimentation or drug screening.
Example Applications of Click Chemistry
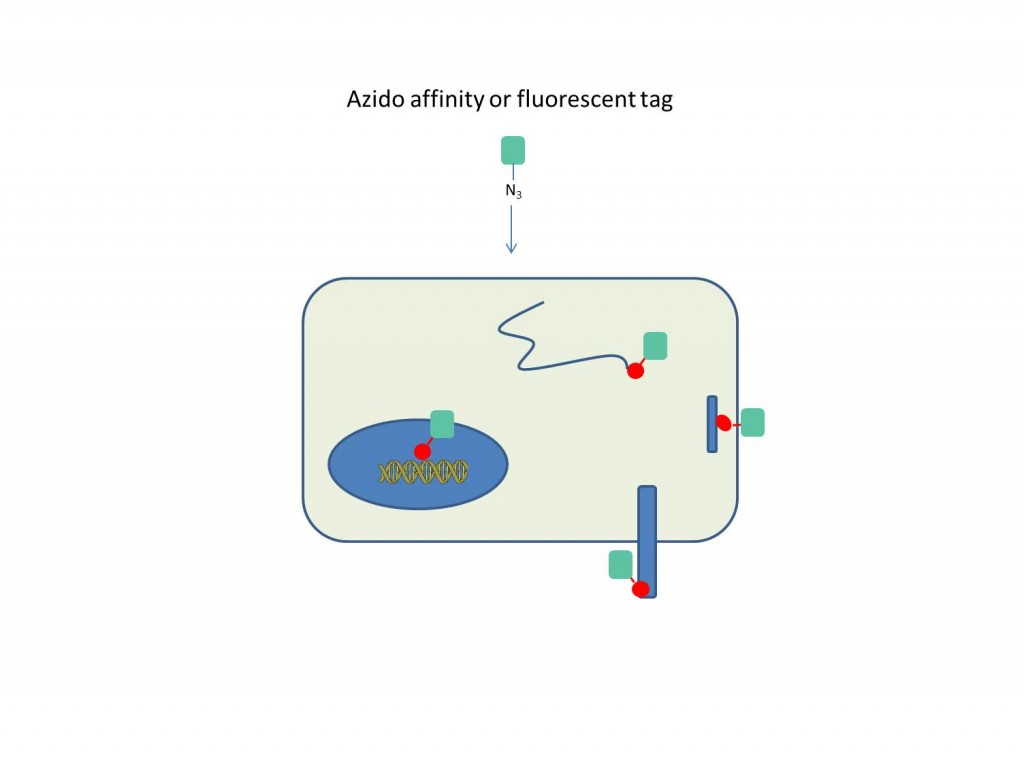
Affinity tagging or fluorescent labeling of biomolecules
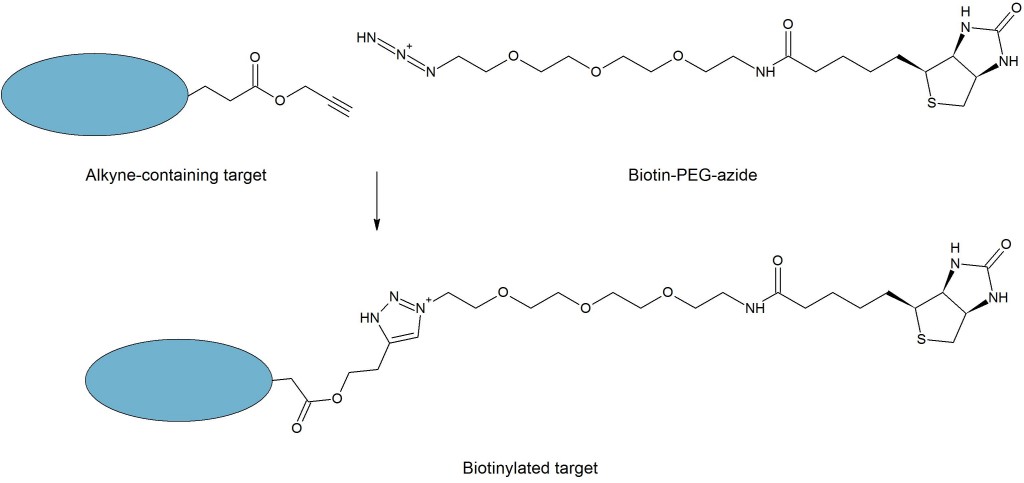
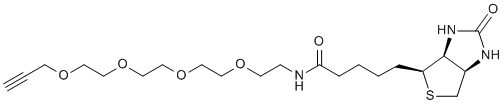
A molecule of interest can be primed for ligation to a variety of affinity or fluorescent tags through direct incorporation of a clickable moiety. A click ligation reaction with a partner containing a fluorophore or affinity tag labels the molecule with the desired tag. Shown is a click ligation linking a biotinylating agent with a ligand via a triazole.
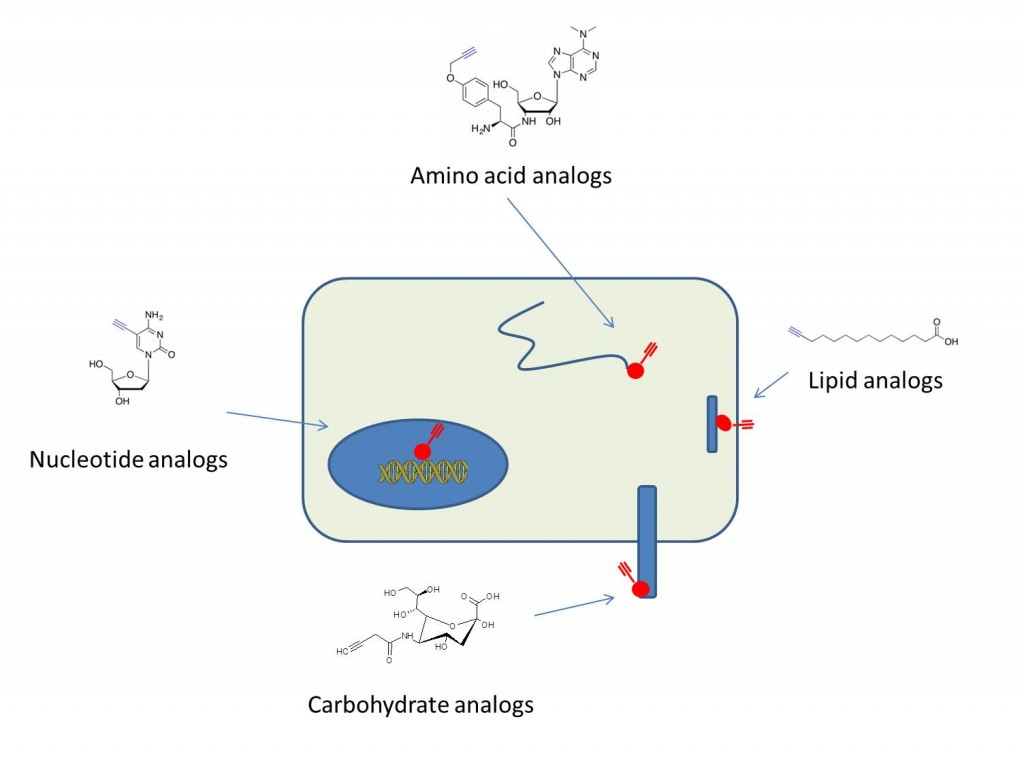
Metabolic labeling
Azides can be readily incorporated into a variety of biomolecules in situ through metabolic labeling. Azide-labeled molecules can then be reacted with an alkyne-labeled fluorophore or affinity tag and visualized or purified accordingly. Separating the metabolic labeling from the detection avoids direct labeling using large affinity tags or fluorophores, which can interfere with biological function or create non-specific effects. Also, it allows kinetic study of cellular processes through pulse-chase labeling experiments. The four examples shown include labeling of the terminal amino acid of nascent peptides using O-propargyl-puromycin(1), fatty acid acylation of protein using alkynyl-myristic acid reporter(2), glycosylated surface proteins using acetylene-sialic acid gycans(3), and DNA with 5-ethynyl-2’-deoxycytosine(4).
| Step 1: Metabolic labeling | Step 2: Addition of tag for detection or purification |
 |
 |
- J Liu et al. Proc. Natl. Acad. Sci. USA 2012 109:413
- N Burnaevskiy et al. Nature 2013 496:106
- ST Laughlin et al. Science 2008 320:664
- L Guan et al. Chembiochem 2011 12:2184
Inquire for clickable tools for metabolic labeling
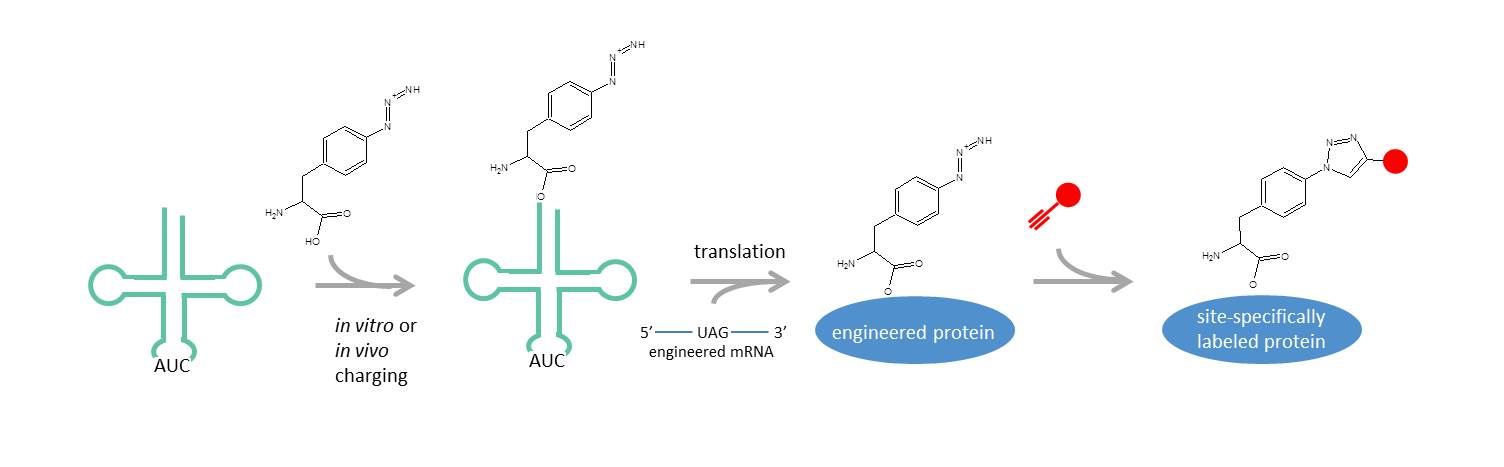
Site-specific protein labeling through incorporation of unnatural amino acids
Clickable amino acids can be incorporated at single sites within recombinant proteins through incorporation of unnatural amino acids. tRNA/tRNA synthetases, typically for the amber stop codon (UAG), are engineered to accept noncanonical, i.e. clickable, amino acids and a corresponding UAG codon is engineered into the desired labeling position in the mRNA encoding the protein of interest. This approach has been used successfully in microbial as well as mammalian cells to generate proteins ready for site-specific labeling(1).
1. CH Kim et al. Curr. Opin. Chem. Biol. 2013 17:412
Inquire about unnatural, clickable amino acids for site-specific protein labeling.
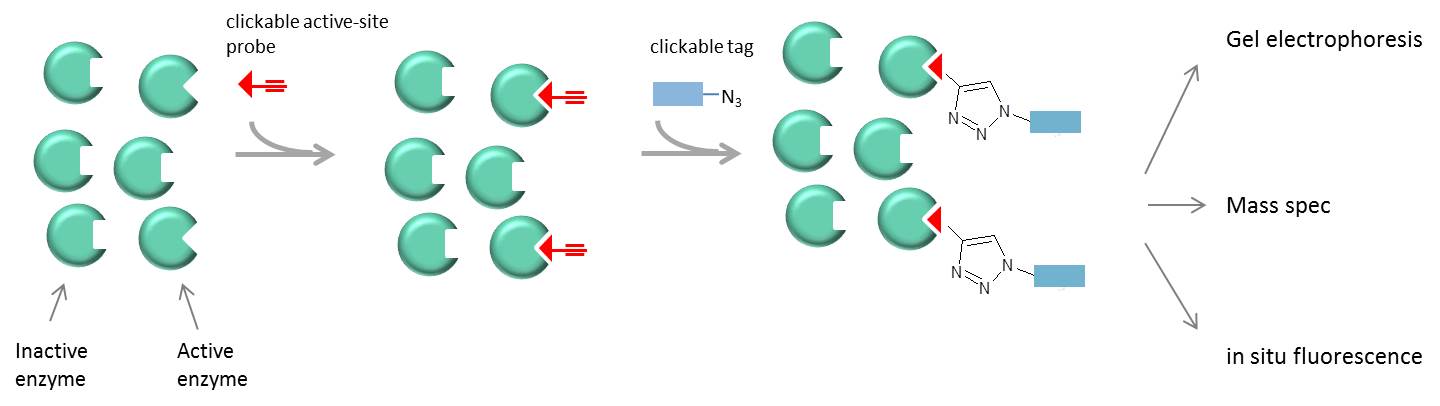
Activity-based protein profiling
Traditional proteomic methods detect proteins based on their abundance rather than their activity. Clickable probes that bind only to active enzymes can be used to monitor enzyme activity in complex samples. These approaches utilize covalent-binding probes, e.g. a substrate or inhibitor-based probe containing a warhead that covalently links to the enzyme active site, and a clickable moiety for detection. The technique can be coupled to gel electrophoresis and MS or in situ fluorescence detection to monitor the localization of enzyme activation. Probes have been generated for proteases, lipases, methylesterases, glycosidases, kinases, phosphatases, cytochrome P450s, histone deacetylases, and many other protein families (1).
1. AS Raghavan et al. Drug Discovery Today 2009 14:178
Inquire about synthesis of clickable active-site probes
Post-translational modification analysis
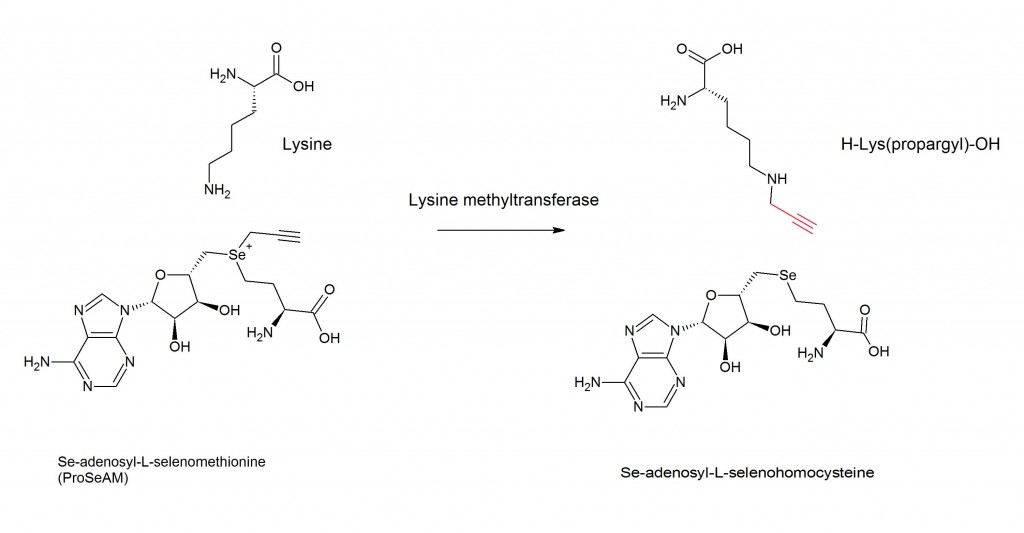
Many post-translational modifications can be monitored using methods similar to the metabolic labeling and activity based protein profiling described above. Clickable analogs of metabolites or substrates of many post-translational modifications, have been developed that can be used metabolically label proteins modified by phosphorylation(1), acetylation(2), methylation(3), glycosylation(4) or sulfenylation(5) with azides or alkynes, which can subsequently clicked to fluorophores or affinity tags for detection/purification. Shown is an example of labeling methylated proteins using a clickable S-adenosylmethionine (SAM) donor (3).
- SE Lee et al. Bioorg. Med. Chem. Lett. 2009 19:3804
- YY Yang et al. J. Am. Chem. Soc. 2010 132:3640
- IR Bothwell et al. J. Am. Chem. Soc. 2012 134:14905
- ST Laughlin et al. Science 2008 320:664
- CE Paulsen et al. Nat. Chem. Biol. 2011 8:57
Inquire about clickable post-translational modification substrates and metabolites
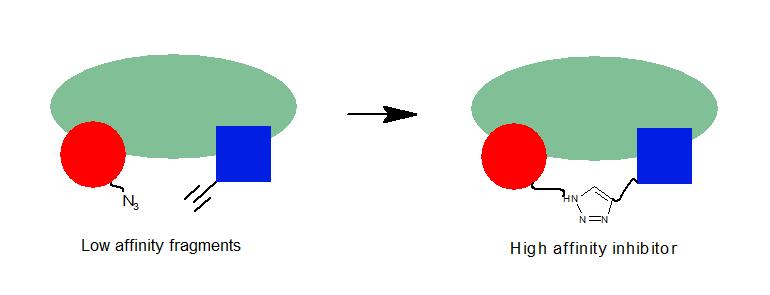
In situ drug discovery
The surface of an enzyme target can be used as a reaction vessel to create high-affinity inhibitors from low affinity fragments. Since the click reaction is catalyzed through proximity, only fragments that bind the target effectively are reacted to form potent multivalent inhibitors. This technique was pioneered by Sharpless to generate a 77fM inhibitor of acetylcholinesterase(1-3).
- R Manetsch et al. J. Am. Chem. Soc. 2004 126:12809
- A Krasinksi et al. J. Am. Chem. Soc. 2005 127:6686
- WG Lewis et al. Angew. Chem., Int. Ed. 2002 41:1053
Med Chem 101 can help you generate the custom small molecule click chemistry reagents needed for your project. Inquire to set up a consultation with our chemists.